
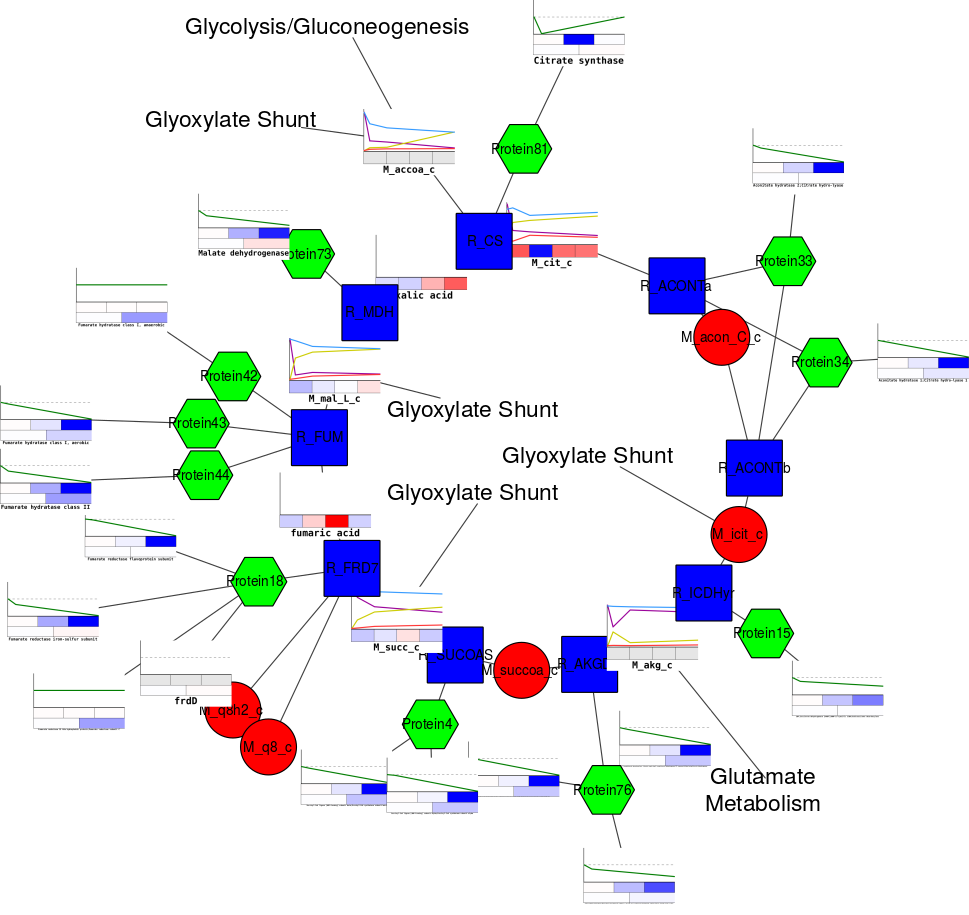
Load molecular and genetic interaction data sets in many formats For example, input a set of custom annotation terms for your proteins, create a set of confidence values for your protein-protein interactions.Ĭytoscape supports many use cases in molecular and systems biology, genomics, and proteomics: Using this feature, you can load and save arbitrary attributes on nodes, edges, and networks.
Delimited text files and MS Excel™ Workbook are also supported and you can import data files, such as expression profiles or GO annotations, generated by other applications or spreadsheet programs. Most of the plugins are freely available.Ĭytoscape supports a lot of standard network and annotation file formats including: SIF (Simple Interaction Format), GML, XGMML, BioPAX, PSI-MI, SBML, OBO, and Gene Association. Plugins may be developed by anyone using the Cytoscape open API based on Java™ technology and plugin community development is encouraged. Plugins are available for network and molecular profiling analyses, new layouts, additional file format support, scripting, and connection with databases. Additional features are available as plugins. Cytoscape core distribution provides a basic set of features for data integration and visualization. Although Cytoscape was originally designed for biological research, now it is a general platform for complex network analysis and visualization.
DOWNLOAD CYTOSCAPE 2.8 SOFTWARE
GeneMANIA is actively developed at the University of Toronto, in the Donnelly Centre for Cellular and Biomolecular Research, in the labs of Gary BaderĪnd Quaid Morris, with input from an independent scientific advisory board.Cytoscape is an open source bioinformatics software platform for visualizing molecular interaction networks and biological pathways and integrating these networks with annotations, gene expression profiles and other state data. Click Ok until you get back to the plugin selection page.
DOWNLOAD CYTOSCAPE 2.8 INSTALL
Note: If you have already installed an official GeneMANIA release, you need to remove it before you can install a development snapshot.


 0 kommentar(er)
0 kommentar(er)
